Scientist Kevin Matthew Byrd, DDS, PhD, founder of the Human Cell Atlas Oral & Craniofacial Bio-network, publishes pre-print article using CellenicsⓇ
The first integrated periodontal meta-atlas of single-cell RNA sequencing data
In their pre-print article, entitled “Evidence for polybacterial intracellular coinfection of epithelial stem cells in periodontitis”, Quinn T. Easter and Kevin Matthew Byrd from the American Dental Association (ADA) Science & Research Institute and their team of collaborators (German Beldorati Stark, Biomage Ltd.; Catherine L. Worth and Brayon Fremin, the Bioinformatics CRO; Kathryn Miller, Akoya Biosciences, Inc.; Mandy Bush and Shannon M. Wallet, the University of North Carolina at Chapel Hill; Blake M. Warner and Janice Lee, the National Institute of Dental and Craniofacial Research; Inês Sequeira, Queen Mary University of London; Jinze Liu, Virginia Commonwealth University; Kang I. Ko, the University of Pennsylvania; and Sarah A. Teichmann, Wellcome Sanger Institute and University of Cambridge) present the first integrated periodontal meta-atlas of single-cell RNA sequencing data using CellenicsⓇ open source software.
Dr. Easter is a Senior Research Associate at and Dr. Byrd is the Senior Manager of the Lab of Oral and Craniofacial Innovation (LOCI) at the ADA Science & Research Institute. Research interests at the LOCI focus on the oral cavity, including biofluids, computational pathology, and precision periodontal medicine. Both Dr. Easter and Dr. Byrd are interested in the identities and spatial relationships of cells in the periodontium, with a particular interest in structural immunity i.e., coordination of specific immune responses in niches. They envision future use of CellenicsⓇ to support more primary and secondary analyses of their single-cell data.
Read their abstract and partial research summary (first posted by Kevin Matthew Byrd on Twitter) below.
Abstract from pre-print article "Evidence for polybacterial intracellular coinfection of epithelial stem cells in periodontitis"
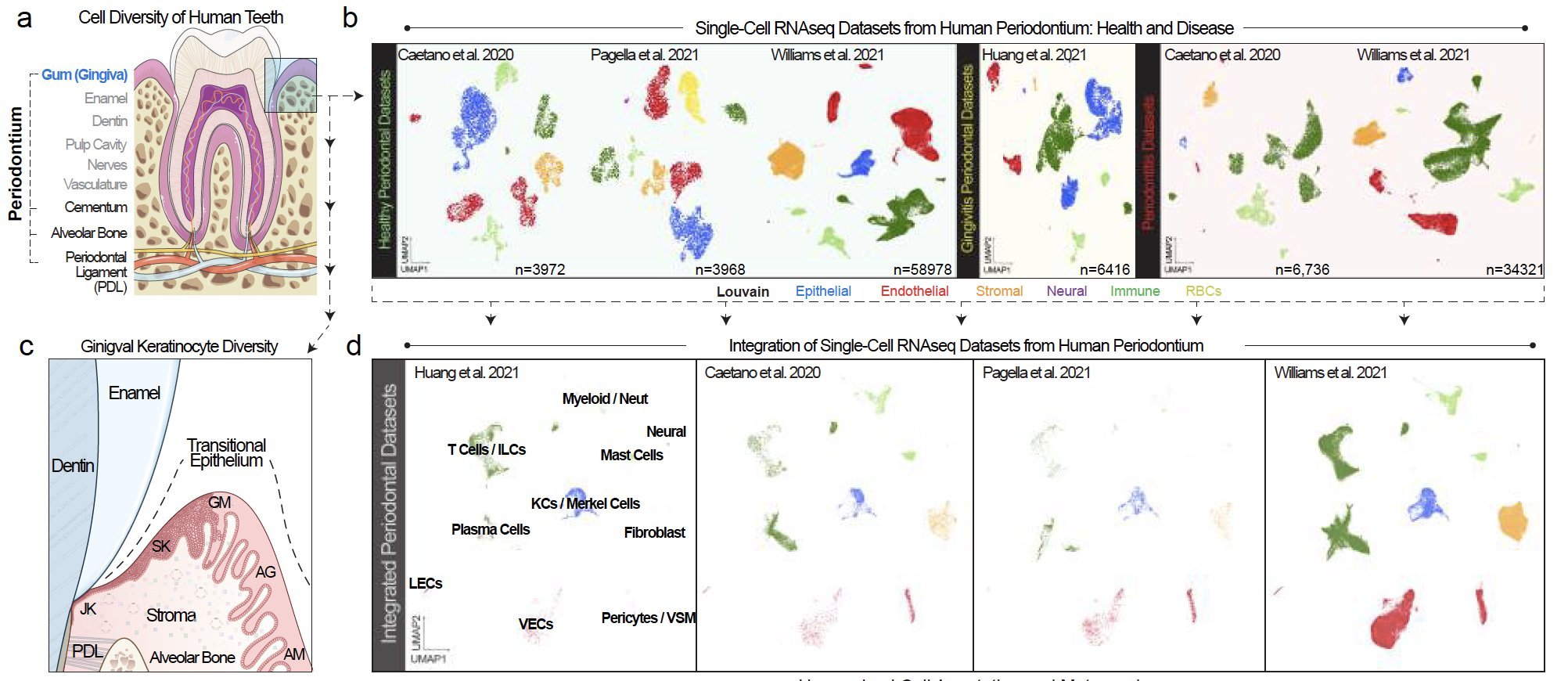
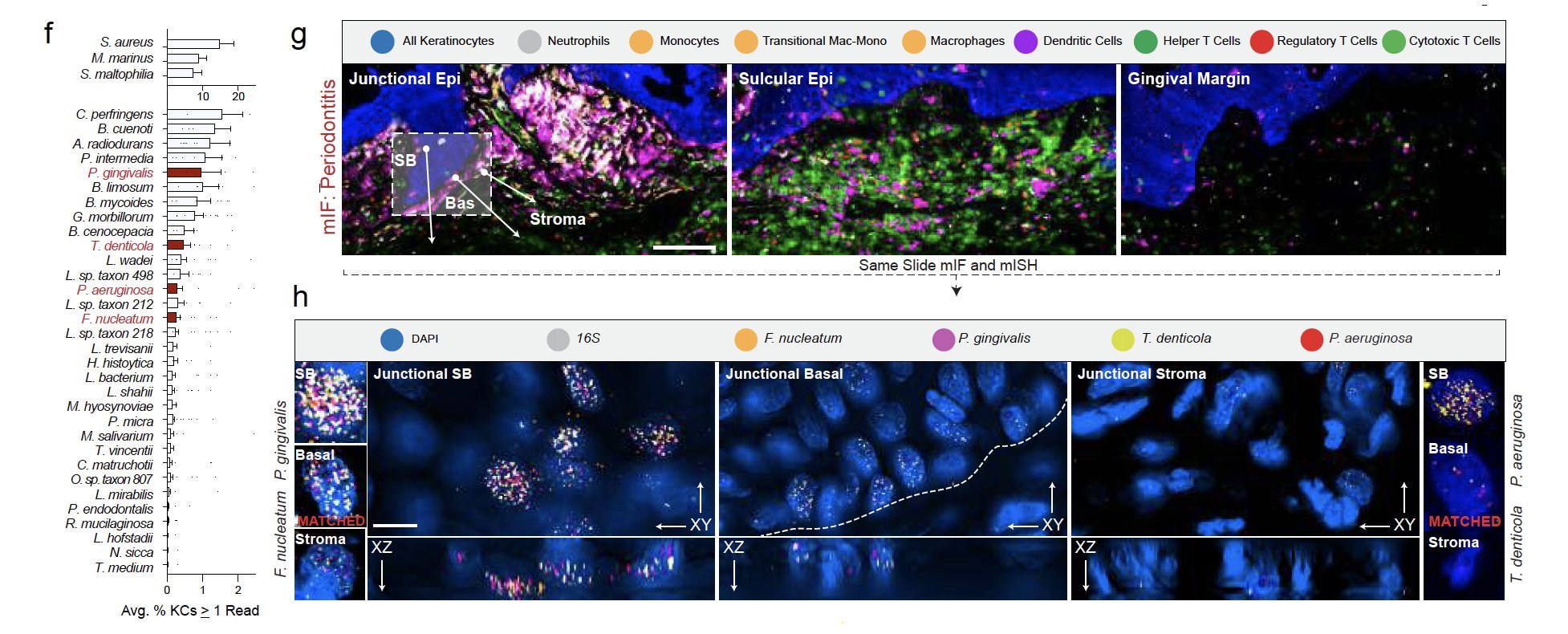
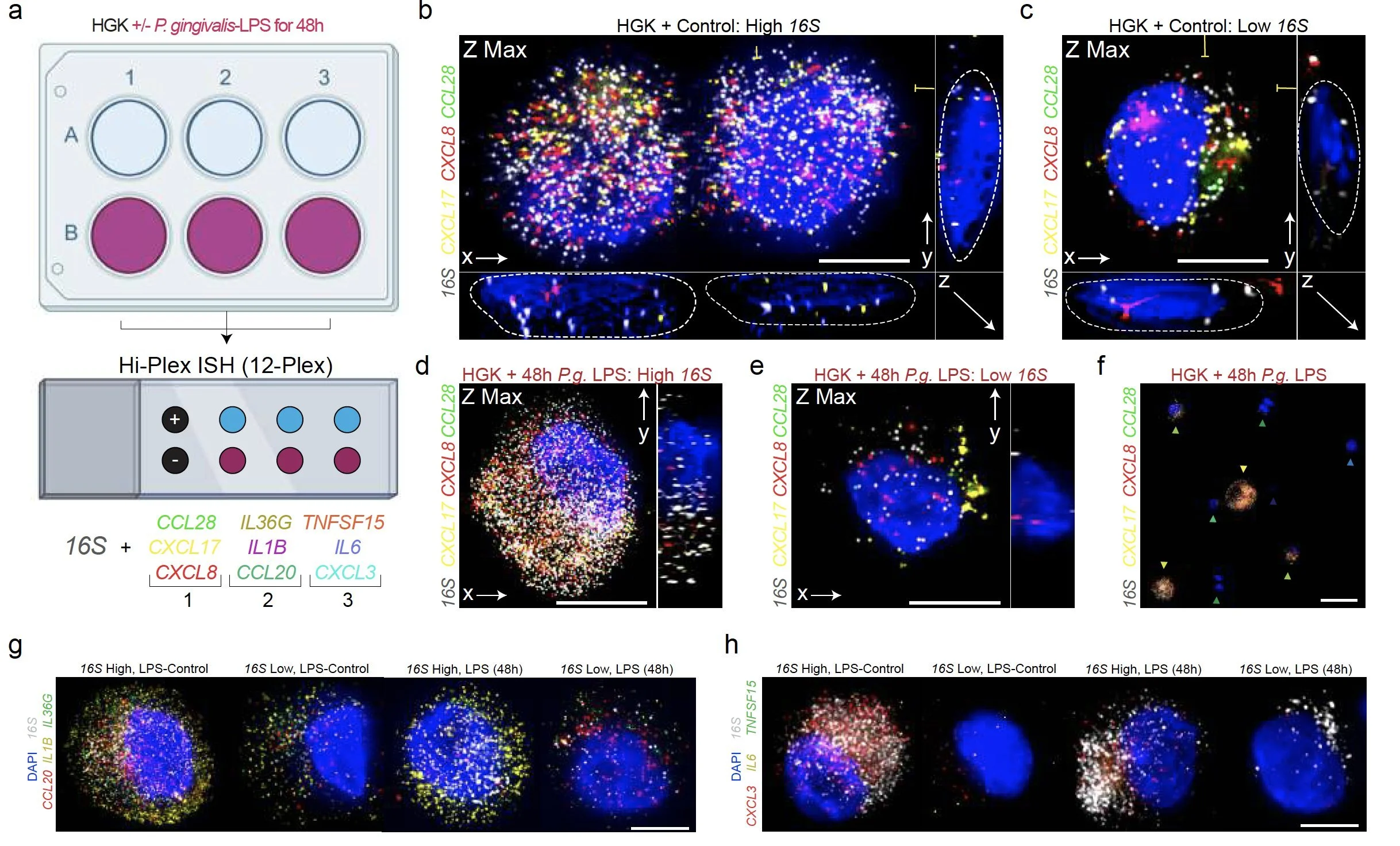
Periodontitis affects billions of people worldwide. To address interkingdom relationships of microbes and niche on periodontitis, we generated the first single-cell meta-atlas of human periodontium (34-sample, 105918-cell), harmonizing 32 annotations across 4 studies. Highly multiplexed immunofluorescence (32-antibody; 113910-cell) revealed spatial innate and adaptive immune foci segregation around tooth-adjacent epithelial cells. Sulcular and junctional keratinocytes (SK/JKs) within epithelia skewed toward proinflammatory phenotypes; diseased JK stem/progenitors displayed altered differentiation states and chemotactic cytokines for innate immune cells. Single-cell metagenomics utilizing unmapped reads revealed 37 bacterial species. 16S and rRNA probes detected polybacterial intracellular pathogenesis (“co-infection”) of 4 species within single cells for the first time in vivo. Challenging coinfected primary human SK/JKs with lipopolysaccharide revealed solitary and synergistic effects. Coinfected single-cell analysis independently displayed proinflammatory phenotypes in situ. Here, we demonstrate the first evidence of polybacterial intracellular pathogenesis in human tissues and cells—potentially influencing chronic diseases at distant sites.
Summary of the research
Periodontitis is an incredibly common disease, but complete niche annotation is still lacking in single cell/spatial approaches. We integrated 4 scRNA-seq datasets to create the first integrated periodontal meta-atlas using open-source software CellenicsⓇ and Cellxgene.
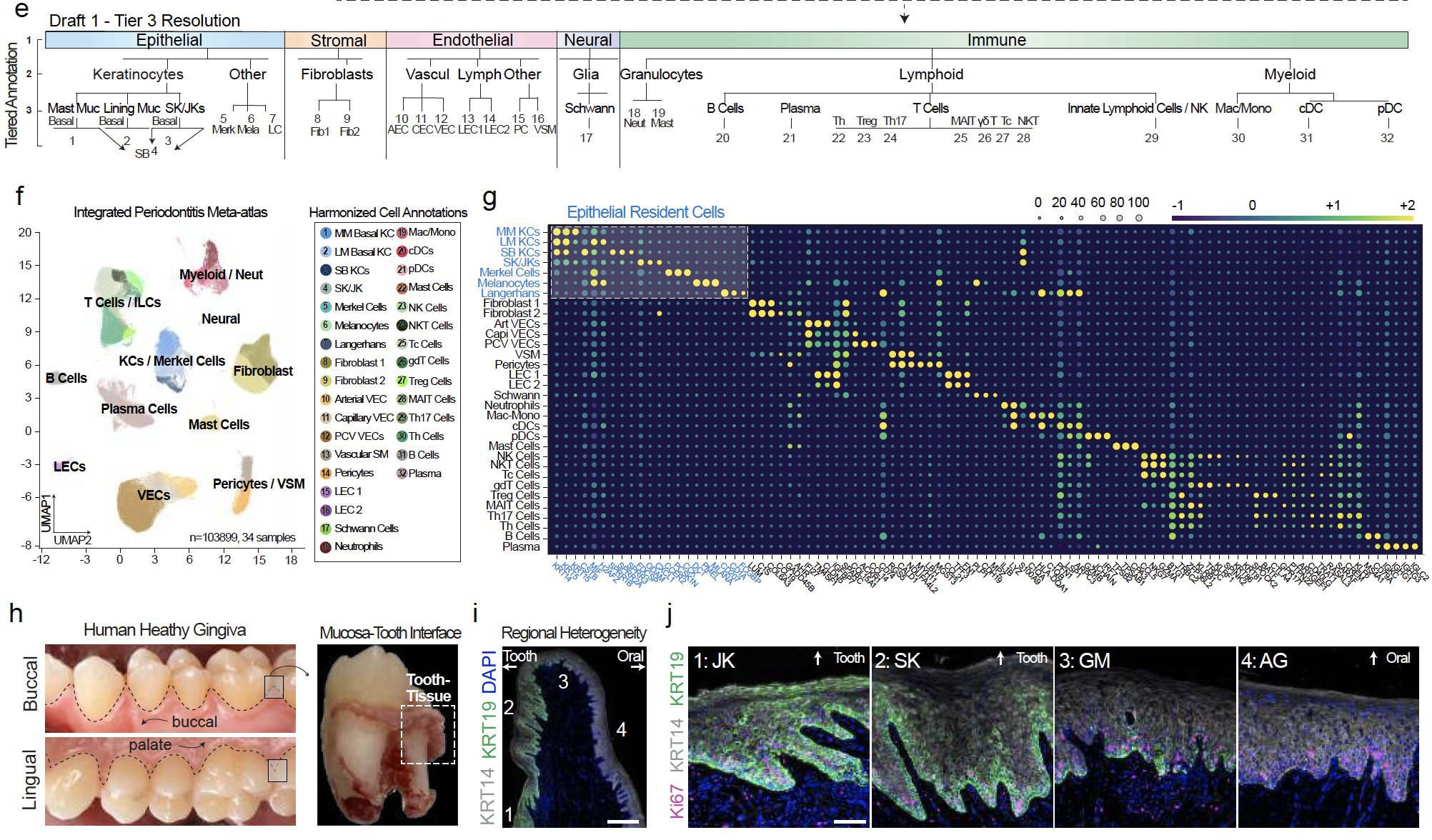
The epithelial attachment to the tooth surface is a unique niche in the human body, but cell type annotation in this transition zone is poorly understood in humans, whether in health or disease. In our analysis, we harmonized annotations for 32 cell types.
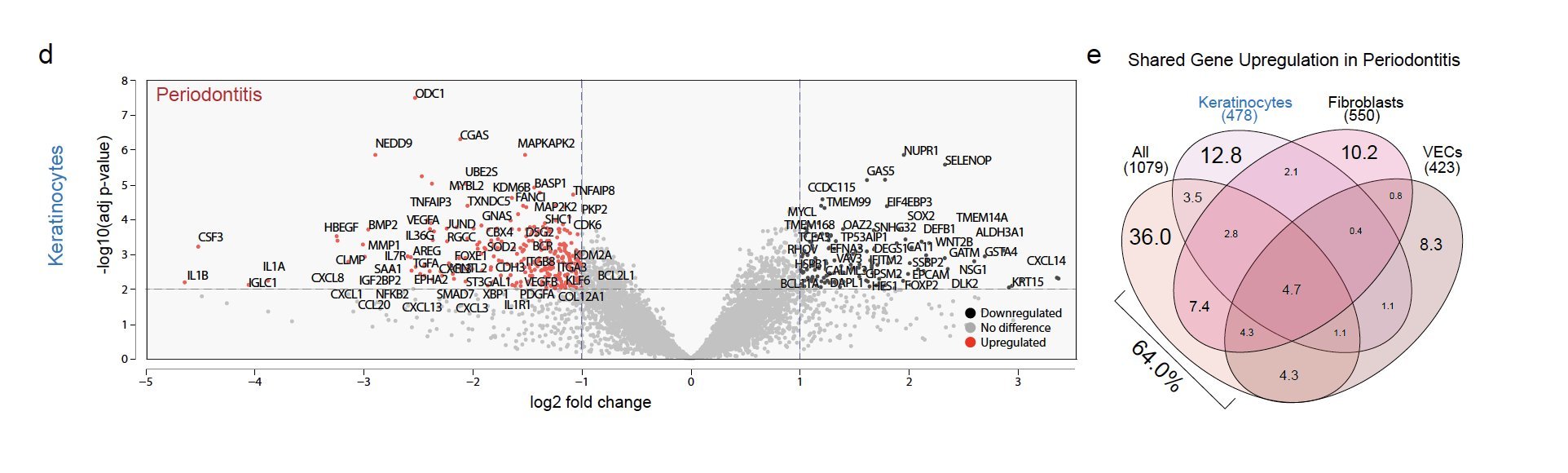
We discovered innate/adaptive immune foci were abutting keratinocytes (KC). We showed that ~2/3 of differentially expressed genes in periodontitis come from just KCs, fibroblasts, and vascular endothelial cells. KCs near the tooth are some of the highest active cell types when assessing receptor-ligand interactions, even in health.
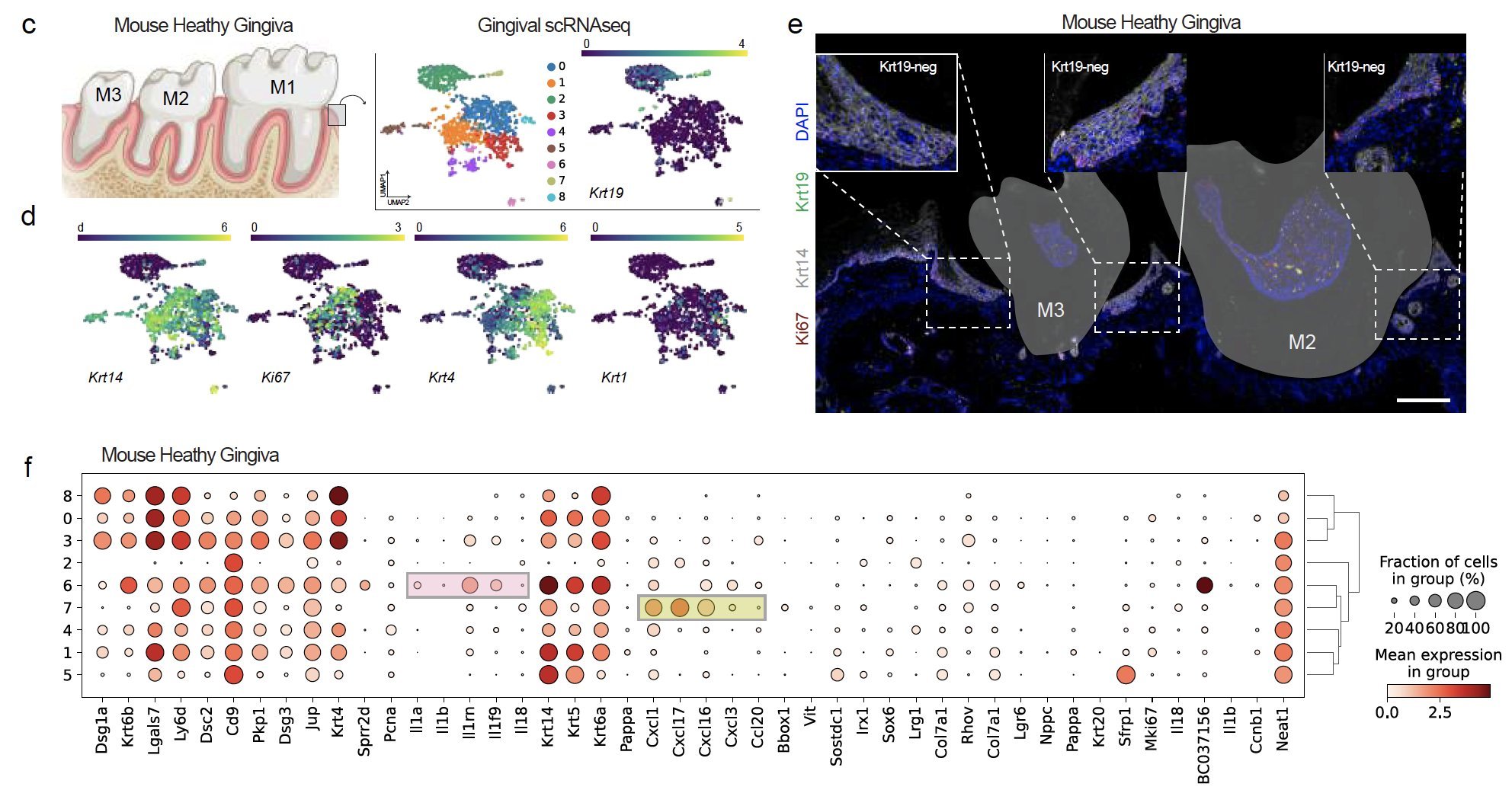
We identified the first signatures of new KCs around the tooth using single cell RNAseq and confirmatory RNA in situ hybridization (ISH) of human tissues. We identified that mice, a commonly used species for periodontitis models, do not reveal the same keratinocyte populations.
In periodontitis, these newly annotated KC altered differentiation signatures that match the upregulation of cytokines that favor innate immune signaling.
We validated bacterial reads in single-cell data by using strict call criteria and ISH to validate cell-specific invasion profiles of well-known 'perio-pathogens' - P. gingivalis, F. nucleatum, P. aeruginosa, and T. denticola. Sometimes all four are found in the same cell, which we saw on the same slide using the Akoya PhenoCycler Fusion and ISH.
We found primary cells from the gingiva can harbor the same phenotypes and periopathogens at the same time. As the cell gets larger (i.e., differentiates in vitro), the cytoplasm grows and so do the number of ISH probes.
We found that polybacterial intracellular coinfection (PIC) is proinflammatory without LPS and only some keratin-expressed cytokines - "keratokines" - synergized with LPS challenge.
Our single-cell metagenomics approach has generated more hypotheses than approached, especially bacteria-driven immunosuppressed phenotypes in disease at a single-cell and spatial context, likely differing across age, sex, and genetic ancestry. Finding bacterial infection in both tissue and primary cell lines asks new questions about current periodontitis treatment effectiveness. Finding new ways to clear intracellular pathogen residency may allow true restoration of the niche to promote health over lifespan.
Disclaimers:
Cellenics® is an open source tool developed by © 2020-2023 President and Fellows of Harvard College.
Quinn, Kevin and colleagues performed their single cell RNA-seq data analysis using the Biomage-hosted instance of Cellenics® that’s available at: https://scp.biomage.net/