Leveraging Public scRNA-seq Data: A Guide to Repositories and Resources
Welcome to the thrilling world of single-cell RNA sequencing (scRNA-seq)! This cutting-edge technology has revolutionized our understanding of gene expression in individual cells, revealing the intricate details of cellular heterogeneity and shedding new light on the mysteries of biology.
But what if you're a researcher who doesn't have the resources needed to conduct your scRNA-seq experiments? Or maybe you want to compare your research to previously published studies? Don't worry - an exciting solution is just a few clicks away. There's an ever-growing wealth of publicly available scRNA-seq datasets waiting to be explored by anyone with an internet connection.
From studies of cancer biology to investigations of neural networks, these datasets cover a wide range of organisms, tissues, and conditions, providing a rich resource for researchers of all backgrounds.
In this blog post, we'll take you through some of the key publicly available scRNA-seq resources, and show you how to start searching for scRNA-seq public data!
scRNA-seq
scRNA-seq is a powerful technology that allows researchers to study gene expression at the level of individual cells. One of the major advantages of scRNA-seq is that it allows us to study cell populations that are otherwise difficult to study, such as rare cell types in cancer. Additionally, scRNA-seq can be used to study the same cells over time, which allows researchers to track changes in gene expression and understand how cells respond to different conditions.
Publicly available datasets
Publicly available scRNA-seq data is a valuable resource for researchers. There are several databases that allow researchers to access and download scRNA-seq data from a variety of species and conditions. You can integrate multiple publicly available scRNA-seq datasets or reuse previously generated data, which can save time and resources that would’ve been spent on setting up your own experiments. With the help of publicly available data, you can also compare your results with previously published data. Additionally, publicly available data can provide a larger sample size, which can be used to draw more accurate conclusions than if you were to rely on a smaller dataset.
Using publicly available scRNA-seq data is like riding a bike on a pre-existing path. You don't have to spend time and energy to create the track yourself, but you can still get the same outcome and even explore new possibilities. You can build on the work that has already been done and use the existing resources to focus on more complex and intricate topics!
Several databases provide access to publicly available scRNA-seq data, including:
The Gene Expression Omnibus
Sequence Read Archive
ArrayExpress
The Single Cell Data Portal
And we are going to tell you more about each of these databases!
Gene Expression Omnibus
The Gene Expression Omnibus (GEO) is a free public data repository provided by the National Center for Biotechnology Information (NCBI) that contains a wide variety of gene expression data, including scRNA-seq data. Data are submitted by the research community! Currently, there are over 4000 datasets available, but note that these are not only scRNA-seq datasets.
GEO DataSets vs GEO Profiles
On the GEO main page, you will notice the options to search at GEO DataSets or GEO Profiles.
In short, GEO DataSets are collections of data that provide a more complete picture of the experimental design and analysis, while GEO Profiles are pre-processed data of individual gene expression profiles that allow for faster and easier comparisons across experiments. Both types of data can be useful depending on the specific research question and analysis approach. However, if you wish to search for scRNA-seq public datasets, GEO DataSets is the right place for you!
Search
GEO allows researchers to search for data by organism, tissue type, and other criteria. To search for gene expression data in the GEO database, go to the GEO website. In the search box at the top of the page, enter a keyword or phrase related to the data you are interested in or an accession ID.
Click on the number of results to view the search results.
Check out the search results and click on the title of the dataset that looks relevant to see more information. As you can see in the example below, the search is by default done at GEO DataSets, as GEO Profiles are focused on individual genes.
Alternatively, click on “Search for Studies at GEO DataSets” on the main page. Then input your keywords or accession IDs into the search box at the top of the page!
Advanced search
There are also advanced search options available on the GEO website that allow you to refine your search based on specific criteria, such as experimental design or data type. These options can be accessed by clicking the "Advanced" button next to the search box on the GEO DataSets page.
You are going to be redirected to GEO DataSets Advanced Search Builder.
Click on “All Fields” to reveal a dropdown of field options. Using one of the available fields will narrow your search results. You can also specify the Boolean operator you want to use in your search.
Here are some explanations for the fields available in the advanced search:
Organism: allows you to select a specific organism or group of organisms to search for data from a particular species or taxonomic group.
Attributes: allows you to search for data based on specific sample attributes, such as age, sex, disease state, experimental factors or treatment condition.
Platform technology type: allows you to select a specific technology type to search for data generated using that technology.
You can also click on “Show index list” to reveal the values for your chosen field. This will reveal specific keywords that you can use to specify your search with that field.
If you are looking for scRNA-seq data, we suggest inputting "Expression profiling by high throughput sequencing" as the DataSet type. You can also select "single cell" as the Sample source.
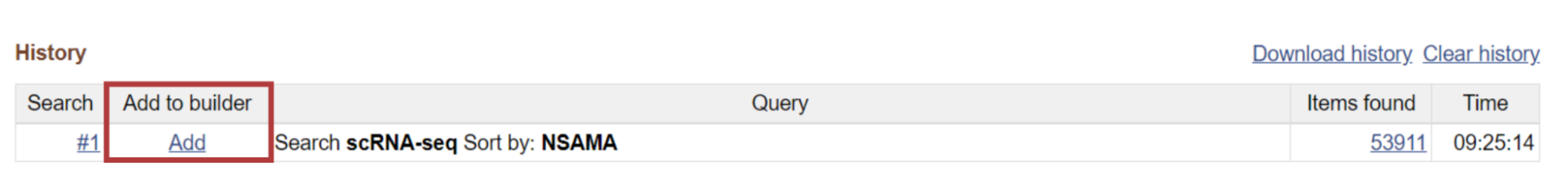
An amazing feature of the Advanced Search Builder is the ability to see your previous searches under the History block.
Moreover, you can add your keywords to the query list in History instead of clicking Search. Just click on “Add to history” next to the search button.
You can also add previously used keywords to a new search (see column Add to builder).
Downloading data
To explore a dataset of interest, click on its title on the search result page.
On the dataset page, you can explore the data by clicking on links to related experiments, samples, and data files. A tip from us – pay attention to the design section of the study you found! scRNA-seq data in GEO may be generated using different platforms, protocols, and quality standards, so it is important to carefully evaluate the data quality and design before using it for your research. For example, it would be important to know if the data has already been normalized or not.
You can download data of interest and various files related to the dataset at the bottom of the dataset page. You would find files that can be directly uploaded into secondary analysis and visualization tools (like Cellenics® open source software) or converted into a compatible file format in the Supplementary file section. Click on the download link for each file that you want to download. You may see two data download options - ftp and http. These data transfer methods overlap in functions, so you can use either one! However, ftp is more suitable for downloading large files, while http could be a better choice for downloading small-sized datasets.
The files may be compressed, so you may need to extract them after downloading. Note that some datasets may have specific terms of use or restrictions on data usage. Additionally, some datasets may be very large, so you may need to have sufficient disk space to download the data files.
Raw data would be available through the Sequence Read Archive, read more about it in the next section.
Sequence Read Archive
The Sequence Read Archive (SRA) is another database provided by NCBI that contains raw sequencing data, including scRNA-seq data. SRA also stores alignment information and annotation files.
Researchers can deposit their sequencing data in the SRA, which makes it available to the scientific community for further analysis and interpretation. The SRA provides a standardized way of storing and sharing sequencing data, which helps to ensure that data is accessible and usable for a wide range of applications.
The SRA is freely accessible to the public, and users can search for data using a variety of criteria, including organism, sequencing platform, and experimental conditions. SRA provides several ways to search for data. Here are some of the most common methods.
Simple search
The SRA website has a search box on the main page that allows you to enter a keyword or accession number to search for data. This is a quick way to find data if you know what you are looking for.
There are four hierarchical levels of SRA accessions:
STUDY (accession forms SRP#, ERP#, or DRP#);
SAMPLE (accession forms SRS#, ERS#, or DRS#);
EXPERIMENT (accession forms SRX#, ERX#, or DRX#);
RUN (accession forms SRR#, ERR#, or DRR#).
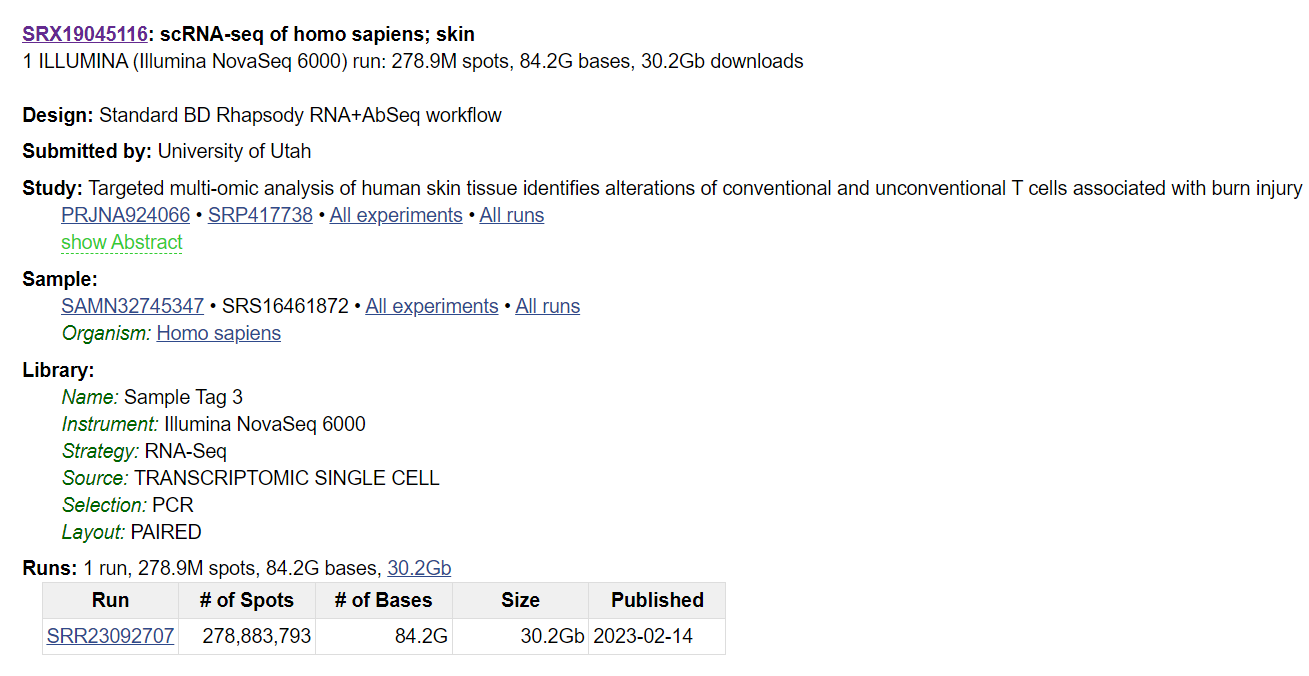
These codes can be useful, for instance, if you want to search for all samples in a study. Click on the sample you have found during your search and find the SRA accession for the study. Then, search using this accession code, which will allow you to see all the samples in the specific study.
You can also modify your search results by including the following terms in your query called Boolean operators (note the uppercase letters):
OR finds records that contain at least one of the terms;
AND finds records that contain all terms;
NOT in front of the search term excludes records matching this term.
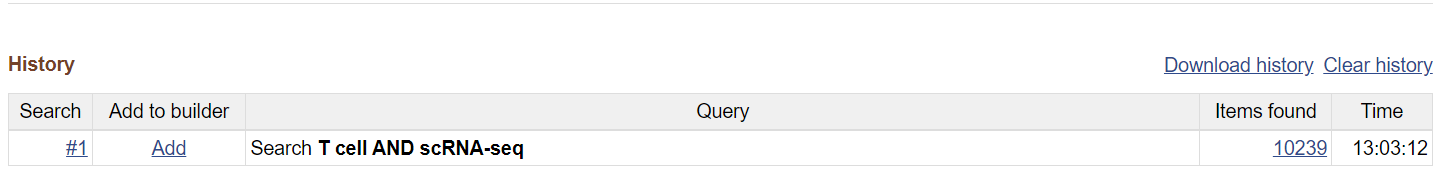
Here is an example of the search result page of a simple search with keywords T cells AND scRNA-seq. You can filter the results further by using the properties on the left-hand side or specifying the taxon on the top right.
Advanced search
The SRA also provides an advanced search page that allows you to search for data using a variety of criteria, such as organism, experimental condition, sequencing platform, and data type. You can combine multiple criteria to narrow down your search.
On the main page, click Advanced. This is going to take you to SRA Advanced Search Builder.
Click on “All Fields” to reveal a dropdown of field options. Using one of the available fields will narrow your search results. You can also specify the Boolean operator you want to use in your search. If you are looking for scRNA-seq data, we suggest specifying “Strategy” field and inputting the keyword RNA-Seq or specifying “Source” field and inputting Transcriptomic single cell.
Click on “Show index list” to reveal the values for your chosen field. For example, here is the index list for the field “Platform”.
Like in GEO Advanced Search Builder, you can see previous searches under the History block.
You can add your keywords to the query list in History instead of clicking Search. You can also add previously used keywords to a new search (see column Add to builder).
Command-line tools
If you prefer to use command-line tools, you can use the NCBI Entrez Direct utilities to search for and download data from the SRA. For example, you can use the "esearch" command to search for data using a query, and the "efetch" command to download the data.
Downloading data
Once you have found the data you are interested in, you can view the metadata associated with the data to learn more about the experimental conditions, sequencing platform, and other relevant information. The metadata can be downloaded straight from the search result page.
To download the data:
Install the SRA Toolkit, which is a command-line tool for accessing and downloading data from the SRA. The SRA Toolkit is compatible with Windows, MacOS, and Linux operating systems.
Open a terminal or command prompt and type the following command to download the SRA data:
fastq-dump --split-files <SRA run accession number>
This command will download the SRA data and convert it into FASTQ format, which is a standard format for storing raw sequencing data.
Depending on the size of the data set and the speed of your internet connection, the download may take several hours or days to complete. Once the download is finished, you should see one or more FASTQ files in the directory where you ran the download command.
You can now use the FASTQ files to perform analysis, such as read mapping.
For example here is the download command prompt for the run SRR23092707 (from the image above).
Keep in mind that downloading large amounts of data from SRA can be time-consuming and may require significant storage space on your local machine or server. You may also need to consult the documentation for the SRA Toolkit or other software tools to optimize your download and analysis pipelines for the specific data set you are working with.
You can also download the data in the Run Browser, however, you can only download one run containing less than 5 Gb of sequence at a time.
ArrayExpress
The ArrayExpress is a functional genomics data collection that stores data from high-throughput genomics experiments, including scRNA-seq data. It is provided by the European Bioinformatics Institute (EBI) and contains data from a wide variety of organisms and conditions. Researchers can use ArrayExpress to search for and download data sets that are relevant to their research, or to submit their own data for public sharing.
ArrayExpress also provides extensive metadata associated with each experiment, which can help researchers to interpret the results of the experiment and understand the experimental design.
Search
To search for gene expression data on ArrayExpress, go to the ArrayExpress homepage. In the search box at the top of the page, type in a keyword or phrase related to your research question. Alternatively, you can enter the accession number.
Experiments and array designs in ArrayExpress have accession numbers in the format of:
E-XXXX-n for experiments
A-XXXX-n for array designs
Then click the "Search" button to initiate the search. Browse the search results to find data sets that are relevant to your research question. You can use the filters on the left-hand side of the page to narrow down the results!
We suggest filtering by File type if you wish to upload the data you have found into the Biomage-hosted community instance of Cellenics® for secondary data analysis and visualization. You can select the file types that are either directly compatible with Cellenics® or can be converted into the accepted file formats. Remember we have posted several tutorials on data conversion here!
Downloading data
When you have found a dataset you are interested in click on the title of a data set to view its details, including the metadata and any available download options.
On the right side of the data set page, you can see a block called “Data files”. You can expand this block by clicking on the << button in the top right corner.
You can view the type of files, their size and format. To download an individual file simply click on the file name! Alternatively, click on the “Download all files” at the bottom of the block.
Single Cell Portal
The Single Cell Portal is a database of scRNA-seq data sets, including data from the Human Cell Atlas and other consortia. It was developed by the Broad Institute. Currently, there are a bit over 500 studies in the Single Cell Data Portal, with 29,614,655 cells analyzed.
Creating an account
To create an account on Single Cell Portal, go to the Single Cell Portal website. Click on the "Sign in" button at the top right corner of the page, and then click on the "Create an account" link below the sign-in form. Fill out the registration form with your personal and institutional information, create a username and password, and confirm the password. You will receive a confirmation email with a link to activate your account. Click on the link to complete the registration process. Once your account is activated, you can sign in to Single Cell Portal to access additional features, such as data upload, analysis, and sharing.
Search
To search for scRNA-seq data in Single Cell Portal based on keywords, use the search box and input study titles, descriptions, authors, or metadata.
You can also search using filters - select an organism of interest, tissue type, disease state, or other relevant filters. Don’t forget to click apply when using filters.
Click on a dataset of interest to view its summary.
Download data
Note that to download data, you have to create an account. Data download is also limited to 0.5 TB per day. To download the experimental files, click on the Download tab on the dataset page.
Here, you can view study files available for download! Either download each file individually or click on Bulk download. Some studies will provide additional means of raw/preprocessed data download on the right-hand side of the dataset page.
Data Repository available in the Biomage-hosted community instance of Cellenics®
Publicly available scRNA-seq data is an important resource for researchers. It allows them to access a large amount of data from a variety of organisms and conditions without the need to generate their own data. One of the key challenges of scRNA-seq is data analysis, as the large amount of data generated by these experiments can be difficult to interpret. You can upload data from the public databases we discussed here into the Biomage-hosted community instance of Cellenics®. This doesn’t require any previous bioinformatics knowledge! Start your analysis here https://scp.biomage.net!
Additionally, you can explore publicly available data in our Dataset Repository! We understand that searching for public data and evaluating its quality can be challenging and overwhelming, so the Biomage Team has curated a repository containing several datasets that can be explored immediately!
To access the public dataset repository, click on the “Get started using one of our example datasets!” button in the Project Details view in the Data Management module.
Alternatively, click on the “Create New Project” button, and then choose ‘Select from Dataset Repository’.
You are going to be redirected to our public dataset repository. To use a dataset, click on the “Explore” button.
The dataset is going to be automatically added to your Project list.
You can also combine your own samples with the data from our dataset repository. If you wish to do so, click “Add samples”.
Upload your samples using the upload pop-up. More details on uploading your own data are available here.
Now, you just have to click on “Process project” and start your analysis! If you have any questions regarding data upload or our public data repository, contact us via the community forum (https://community.biomage.net/).