The Human Cell Atlas and Beyond: An Introduction to Single-Cell Data Atlases
Single-cell RNA sequencing (scRNA-seq) has revolutionized our understanding of cellular heterogeneity, providing a powerful tool to unravel the complexity of biological systems at unprecedented resolution. As scRNA-seq technology continues to advance, researchers are generating vast amounts of data, necessitating innovative approaches to analyze, interpret and store this information effectively. One such approach gaining prominence is the use of single-cell data atlases. By developing these atlases, we will be able to improve our understanding of the cellular landscape and develop treatments tailored to each patient's condition.
Atlases provide a comprehensive view of gene expression patterns in cells within a particular organ or tissue. These atlases usually are projects generated through collaborative efforts between multiple research groups and institutions. They can be used to identify new cell types, understand cell-type-specific gene expression patterns, and help to understand the organization and function of cells within different tissues. In this post, we wanted to highlight some of the key atlases containing scRNA-seq data!
What are single-cell data atlases?
A single-cell data atlas is a comprehensive collection of single-cell transcriptomic data from various tissues or organisms, where each cell's gene expression profile is recorded. These atlases allow researchers to navigate and explore the heterogeneity of cellular populations within specific tissues or developmental stages, providing a rich resource for understanding cellular diversity and dynamics.
These comprehensive resources serve as invaluable references for researchers worldwide, enabling previously unimaginable discoveries. As we continue to refine and expand these atlases, we will unlock even greater insights into the intricate world of cells, paving the way for novel therapeutic strategies and personalized medicine.
The Human Cell Atlas
The Human Cell Atlas (HCA) is a global initiative founded in 2016. At its core, the HCA seeks to provide a detailed blueprint of the cellular composition of the human body. The HCA employs cutting-edge single-cell and spatial analysis methods to provide a comprehensive view of the cells in the human body, including their location, function, and gene expression patterns.
Cells are fundamental units of living organisms, and to study human biology we must study human cells. Thus, the HCA is an exceptional resource for studying human biology – both in health and disease. The cellular reference maps generated in HCA allow researchers to discover what governs the differentiation and activity of different cell types, how these cells interact with each other and where they are located within tissues and the body. Then, researchers can study the biological changes that occur in disease and the therapeutic potential of these changes.
Adapted from Lindeboom et al., 2021. Towards a Human Cell Atlas: Taking Notes from the Past.
The data on cells of the human body is available through the HCA Data Portal.
You can explore individual projects in the HCA, and download the available files for your use!
The Cancer Genome Atlas
The Cancer Genome Atlas (TCGA) is another example of a publicly available data atlas. TCGA is a large-scale, collaborative effort between the National Cancer Institute (NCI) and the National Human Genome Research Institute (NHGRI) to generate and analyze genomic data from various types of cancer. The goal of TCGA was to identify new cancer-associated genes and pathways and to understand the molecular basis of cancer in order to improve the diagnosis and treatment of cancer.
TCGA provides a wealth of publicly available data, including scRNA-seq data, on different types of cancer, such as lung, breast, ovarian and many more. Data from TCGA can be used to identify new cancer-associated genes and pathways, and to understand the molecular basis of cancer, helping to improve the diagnosis and treatment of cancer.
The TCGA data is available through the Genomic Data Commons (GDC) Data Portal, which provides a user-friendly interface for searching and downloading the data. However, take note that many projects contain controlled access data and require NIH eRA Commons account and authorization to access the data through the NIH database of Genotypes and Phenotypes.
Other atlases
HCA and TCGA are well-known and influential data atlases. However, there are other smaller-scale atlases that we find interesting and want to highlight!
Mouse Cell Atlas
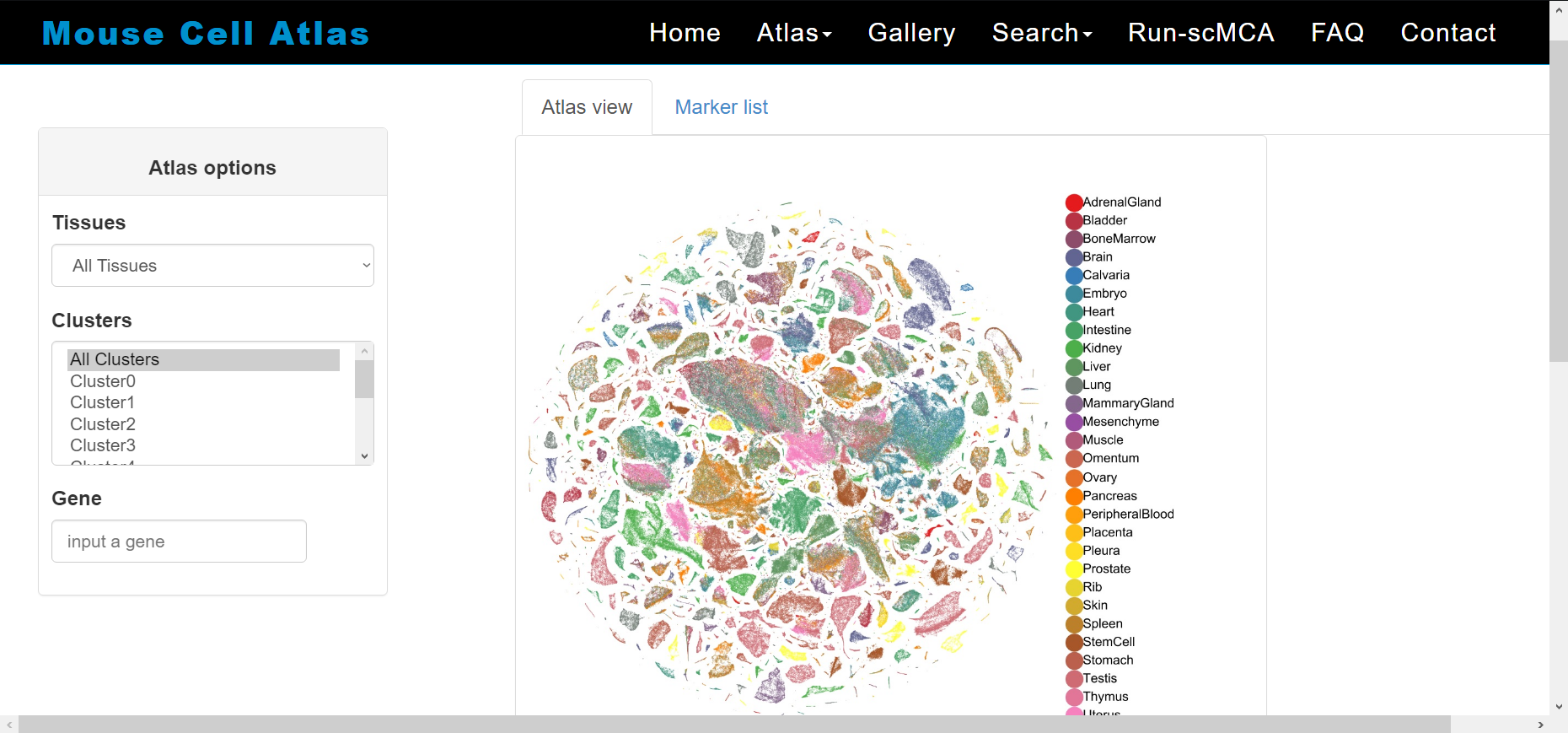
The Mouse Cell Atlas (MCA) is an effort to determine the cell type composition of major mouse tissues using scRNA-seq.
Mapping the Mouse Cell Atlas by Microwell-Seq. Available at https://www.sciencedirect.com/science/article/pii/S0092867418301168
There are several versions of MCA:
In MCA 1.0. researchers analysed >400,000 single cells from >10 mouse tissues
In MCA 2.0 researchers analysed >520,000 single cells at seven mice life stages from the early embryonic stage to the mature adult stage
In MCA 3.0 researchers analysed ~1,130,000 single cells at ten mice life stages
Each version of the MCA is available for exploration on the MCA resource website. Data can also be downloaded for each version.
Fly Cell Atlas
The fruit fly Drosophila is one of the most commonly used model organisms used in biomedical science. The fruit fly is a great model for not only studying fundamental genetics, but disease development as well due to the malleable DNA code of the organism. Single-cell methods have allowed the characterisation of different cell types in the fruit fly and generated comprehensive cellular reference maps.
The goal of the Fly Cell Atlas is to bring together researchers studying Drosophila to build cell atlases of the fruit fly in health and disease using single-cell genomics, transcriptomics, and epigenomics. Currently, the data in the Fly Cell Atlas has been mainly generated using single-nucleus RNA sequencing and is available for public download. You can read more about how the atlas was generated here: https://www.science.org/stoken/author-tokens/ST-363/full!
Figure from Li et al., 2022. Fly Cell Atlas: A single-nucleus transcriptomic atlas of the adult fruit fly
A final note
The landscape of single-cell data atlases is continually expanding, offering researchers a wealth of information to explore and analyze. Atlases, such as the Human Cell Atlas, The Cancer Genome Atlas, and many others, provide a deeper understanding of cellular diversity, developmental processes, and disease mechanisms. Through single-cell data atlases, scientists can now identify and classify rare or previously unknown cell types, uncover cellular heterogeneity within populations, and decipher the molecular mechanisms driving cellular behavior. These atlases act as roadmaps, allowing researchers to navigate the diverse cellular landscapes with precision and uncover critical insights into human health and disease. By leveraging the power of scRNA-seq and other cutting-edge technologies, these atlases are driving scientific discovery and opening up new avenues for personalized medicine, drug development, and therapeutic interventions.
While these data atlases have already made significant contributions to scientific knowledge, they are continually evolving. As technologies advance, atlases will expand to include more tissues, species, and disease contexts, providing a deeper understanding of cellular biology.
In addition to the wealth of knowledge they provide, single-cell data atlases offer researchers the opportunity to explore and analyze the data themselves. Many of these atlases are freely accessible online, allowing scientists to access, download, and utilize vast amounts of single-cell transcriptomic data. The scRNA-seq data from single-cell atlases can be analyzed using Cellenics® - an open-source and cloud-based scRNA-seq analysis software that allows you to analyze your dataset without prior programming knowledge. Biomage host a community instance of Cellenics® that’s freely available for academic users with datasets of <500,000 cells (log in at https://scp.biomage.net/)!